In a joint effort by the Brown University and the University of Wisconsin-Madison, researchers developed an improved 3D imaging method to better understand the epithelial-mesenchymal transition in cells which is implicated in cancer spread, or metastasis. This method possesses potential as a screening tool in providing personal, patient-derived cancer treatment.
In this study published in March, the researchers used the new imaging technique to measure forces generated by cell clusters at a scale not previously possible. “We have a technique now through which we can measure multicellular generated displacement patterns and characterize them for different phenotype conditions. And we can do this on a high throughput scale,” said Mohak Patel, co-first author of the study and a former PhD student in the Wong Lab. Assistant Professor of Engineering and Molecular Pharmacology, Physiology and Biotechnology Ian Wong was the principal investigator of the study.
In order to create this imaging technique and account for the varying degrees of force present in this cell transition process, the researchers conducted a case study. They wanted to understand where and how the epithelial-mesenchymal transition (EMT) occurs in three-dimensional cell clusters. EMT is a process in which epithelial cells, spherical cells that are compact and form clusters, become mesenchymal cells, which are elongated migratory cells that break away, Patel said. In cancer metastasis, one way cancer cells escape primary tumor sites is by undergoing this shift, he added.
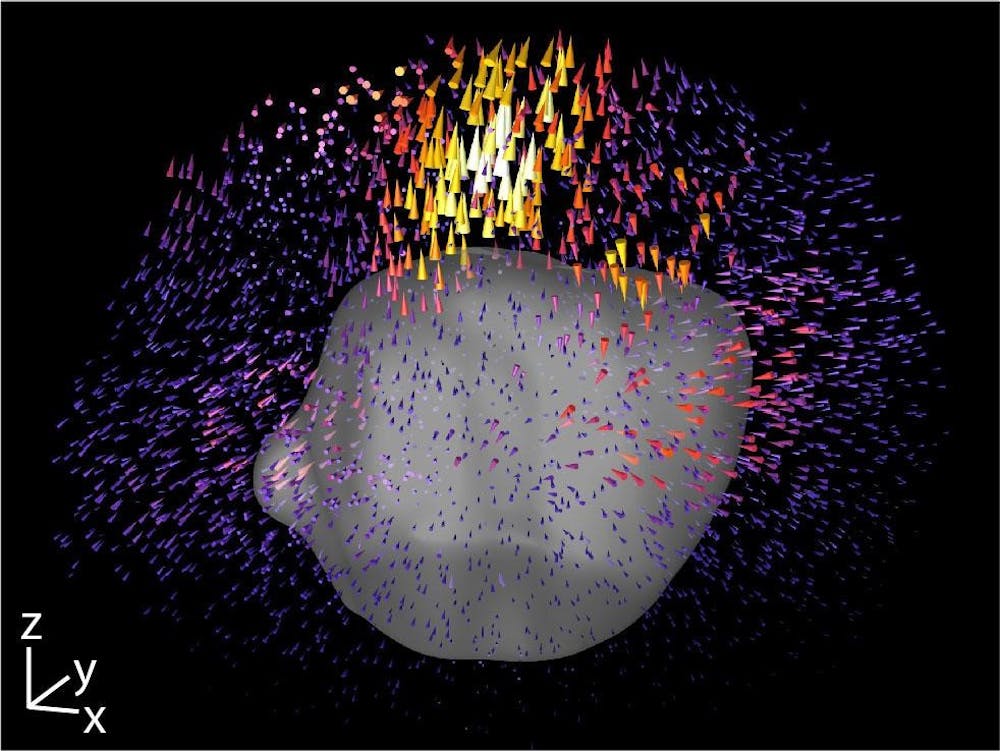
To measure the forces that cell clusters apply in the context of EMT, the researchers used a common technique called traction force microscopy, Patel said. Cells are placed in a silk-collagen hydrogel culture designed to mimic the natural extracellular matrix, which surrounds cells in their 3D microenvironment, said Susan Leggett, co-first author of the study, former PhD student in the Wong Lab and current postdoctoral student at Princeton. Researchers can place beads in this matrix culture and track them to analyze the pulling contractile and pushing protrusive forces exerted by the cells.
Previous traditional analyses using techniques like TFM have been done at the single-cell level, Leggett said. Earlier techniques also spatially averaged the displacement of the cells into a single value, which only works for very few data points or for single-cell analysis. But since the researchers needed a method to study multicellular clusters that are highly variable and have a nonuniform distribution of forces, they developed a new platform called Displacement Arrays of Rendered Tractions, aptly referred to as DART, that extended the use of TFM. With this new method, they could study their 3D clusters effectively.
DART is “a visualization and quantitative tool to understand what the (cell) displacements look like,” Leggett said. Instead of averaging values, researchers broke up the region around the cluster of cells into 16 subvolumes. Next, they looked at each subvolume independently and analyzed the behavior in each region as a function of its spatial localization. Each 3D volume was then flattened into a 2D, easy-to-read map of forces that resembles a dart board, she said. This process allowed the researchers to take data that is very qualitative and create a quantitative metric.
Patel led the team in using this data to train machine learning algorithms to recognize cellular conditions. The researchers found distinct force signatures in EMT that allowed them to classify clusters of cells as epithelial, mesenchymal or in an intermediate state, Leggett said.
Moving forward, this technique could be modified to better understand mechanical forces in other situations in which cells move in an extracellular matrix, such as in tissue development. “An exciting future direction would be the application of DART as a screening tool with patient-derived clinical samples,” Leggett said. Doctors could one day take a sample from patients and test them with different drugs on well plates to see the effects on the cells’ mechanical properties leading to very personalized drug treatments.
“The last decade of work in cancer and other diseases have only begun to shed light on how important mechanical forces are in various physiologic and pathologic processes,” said William Polacheck, assistant professor of biomedical engineering at the University of North Carolina at Chapel Hill, who was not involved in the study. But researchers have lacked an applicable method for high throughput analysis of the mechanical interactions in large cell clusters that comprise tumors.
“We don’t do a lot of measuring of forces, but not because we don’t want to, but because we don’t have the appropriate techniques, which is why I’m excited by this work,” Polacheck said about his lab. The code for the technique used in this study can also be accessed for free online, according to a University press release. Polacheck looks forward to potentially applying the “highly important method” from this study to his own research.
“Approaches like this, ones that can be done with relatively little overhead in lots of labs in the country and world, is a really important advance to allow us to better characterize these forces to start to learn more,” Polacheck said.

Jared is a Senior Staff Writer for Science and Research. He is a senior from Albuquerque, New Mexico studying physiology and biotechnology. Outside of The Herald he likes to fish, ride bikes and research the role of metals in human health and disease.