University researchers have developed a novel imaging technique that distinguishes cells that have gone through a change that occurs during cancerous growth — among other processes — from cells that have not. A study published Oct. 3 describes the development and impressive accuracy of this tool, “the first of its kind to do a really extensive morphological study,” according to Susan Leggett GS, the lead author of the paper.
The approach is preliminary, and “there is a lot of work to be done,” Leggett said, adding that at this stage of development the technique is most powerful as a method for screening new drugs. In the future the tool may be applicable to cancer screenings.
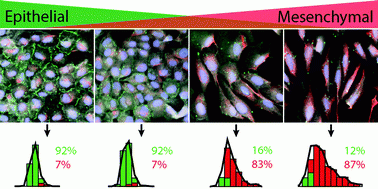
The change that occurs in processes such as wound healing, development and cancerous growth is called the epithelial-mesenchymal transition. Epithelial cells have a cube-like form, while mesenchymal cells are more spindle-shaped, but the cells are different in more than just their shape. The researchers also stained their samples for specific biomarkers — images included in the study show different colors for E-cadherin and vimentin. E-cadherin has higher concentrations in epithelial cells while more vimentin is present in mesenchymal cells.
The researchers used MATLAB to “analyze data on a more macroscopic scale and figure out what the trends were and then train the computer to recognize the transition,” said Zachary Neronha ’19, a co-author of the study. Many different visual characteristics of the cells were observed and recorded, but only the four most predictive factors were included in the final algorithm, Leggett said.
One reason for the elongated form of mesenchymal cells is motility, Leggett said. Epithelial cells can stick together and create tumors, but they will not split apart. If a cell becomes mesenchymal, it is able to break away and form other tumors throughout the body. The numbers of epithelial and mesenchymal cells in a tissue sample can indicate the aggressiveness of a tumor, so this technique could potentially be applied to understanding and predicting cancerous growth.
This new form of single-cell analysis is a step toward replacing human identification of epithelial and mesenchymal cells, providing a more exact judgment of a sample. “Single-cell analyses are a new trend in biology,” Leggett said. “All of this historically has been done by eye, and now we’re finally reaching this vision for biology where we can start to integrate all of this really complex data that we can’t even really wrap our minds around.”
“Single-cell image analysis has been of growing emphasis in cell biology” for five years or so, wrote Douglas Lauffenburger, professor of chemical engineering and biological engineering at the Massachusetts Institute of Technology, in an email to The Herald. But this application is new, he added.
Leggett hopes that through understanding the details of this process on such a small scale, scientists will be able to shed light on how to shift cells back to their epithelial forms. The lab hopes to work on three-dimensional screening and using morphological analysis in individualized medicine in the near future.
This new imaging tool is a “very exciting advance” that holds great promise, Lauffenburger wrote. As the replacement of human judgment with machine analysis increases in biology, “the key will be to complement machine learning analyses with human intuition to provide conceptual explanations of biological processes from the mathematical results,” he added.