A study led by University researchers has pinpointed a mechanism behind some forms of autism, a finding that could guide the development of drugs targeting its underlying cause, said Eric Morrow, assistant professor of biology and psychiatry and human behavior and senior author on the paper. The research, which was motivated by past human studies and conducted on mice, was published earlier this month in the journal Neuron.
The study was a four-year effort inspired by previous research to identify genes involved in autism and related developmental disorders, Morrow said.
Defects in the gene that produces the protein NHE6 were previously known to cause a rare, autism-like disorder called Christianson syndrome. Morrow and collaborators found similar genetic abnormalities in some patients with autism. In January, his team published a paper showing that NHE6 levels are lower in the brains of autistic patients, while levels of a protein related to NHE6 are elevated.
“That shows that this genetic pathway is very important” because it is a “common pathway” between Christianson syndrome and autism, Morrow said.
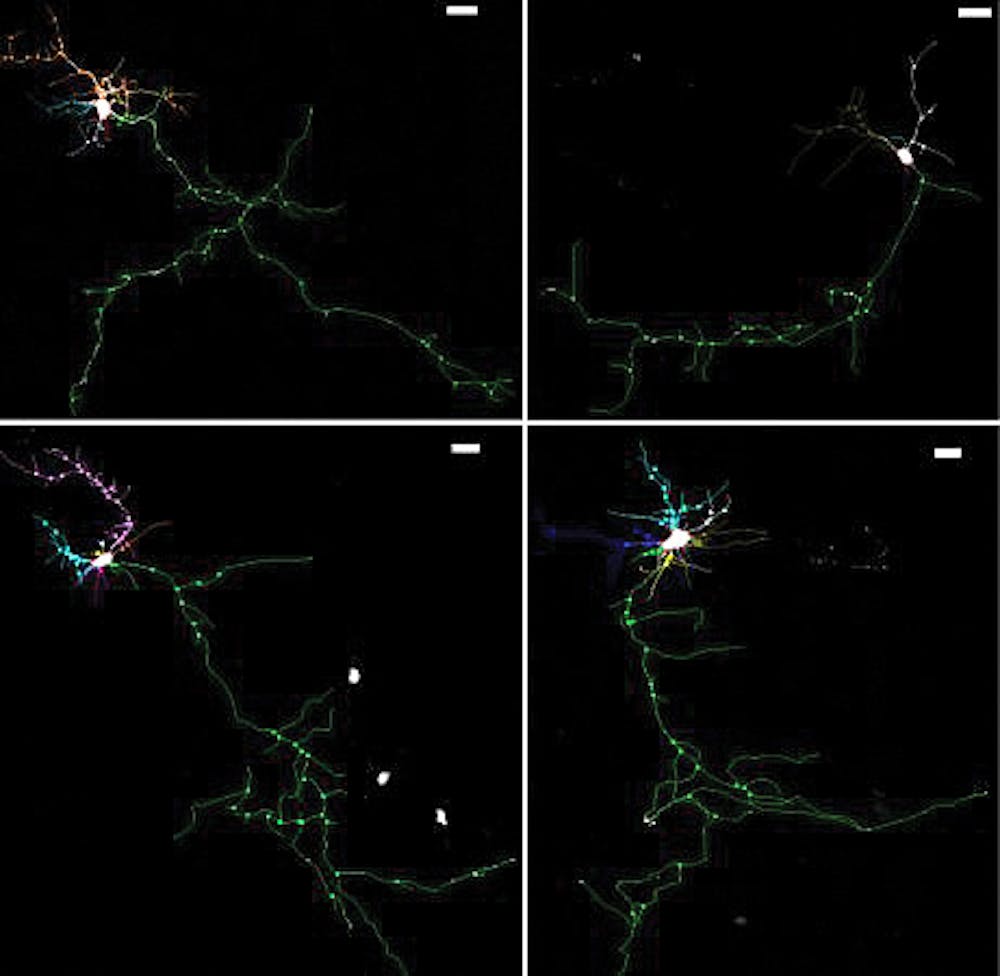
Having identified NHE6 as involved in some forms of autism, Morrow’s team sought to investigate its role in brain development by using mice with non-functional copies of the gene for NHE6. They found brain cells in the mice were unable to form neural branches properly, which is neurologically problematic because branching allows the brain to form circuits, Morrow said.
NHE6 belongs to the family of genes regulating the acidity of endosomes, which are shuttles capable of either recycling or destroying proteins inside cells, Morrow said. The team found that mice without NHE6 had unusually acidic endosomes that disrupted the balance between recycling and destruction, he added.
The authors attempted to restore the balance between the two pathways using a regulatory molecule called BDNF. The treated neurons “grow branches, and they look almost like the wild type,” Morrow said, a striking improvement over the stunted, untreated neurons.
The team’s work represents an advance in our understanding of how genetic defects can contribute to neurological disorders, wrote Sven van Ijzendoorn, associate professor of cell biology at the University of Groningen in the Netherlands, in an email to The Herald. “The function of NHE6 as an endosomal pH regulator was known, but not the physiological relevance,” he wrote. “This paper now shows how NHE6-regulated endosome pH governs neuronal” branching and “how loss of this function gives rise to a human brain disorder,” he added.
Morrow cautioned that while the findings suggest a potential drug mechanism against autism, much work remains even in the ideal case.
This “was a study in a dish,” he said, adding that mouse brains could have “conceivable differences” from humans in ways relevant to the mechanism. It is also challenging to design drugs that can be delivered “in the right place at the right time” and act specifically enough not to cause side effects, he said.
The “good news is we have the tools for these experiments,” he said, such as live mice and ways of growing human brain cells in a dish. He also said compounds similar to BDNF have already been tested in humans and found to be safe.
For Morrow, the immediate goal is not to cure autism. The BDNF molecule might only fix some of the problems caused by the NHE6 defect, he said. But in severe cases where patients are unable to speak or have frequent seizures, even incremental improvements could make a major difference.
ADVERTISEMENT
More




